Investigation of Neolamarckia cadamba Phytoconstituents Against SARS-CoV-2 3CL Pro: An In-Silico Approach
Published 2023-04-12
Keywords
- 3CL Pro; Molecular docking, Neolamarckia cadamba; Phytoconstituents; SARS-CoV-2
How to Cite
Abstract
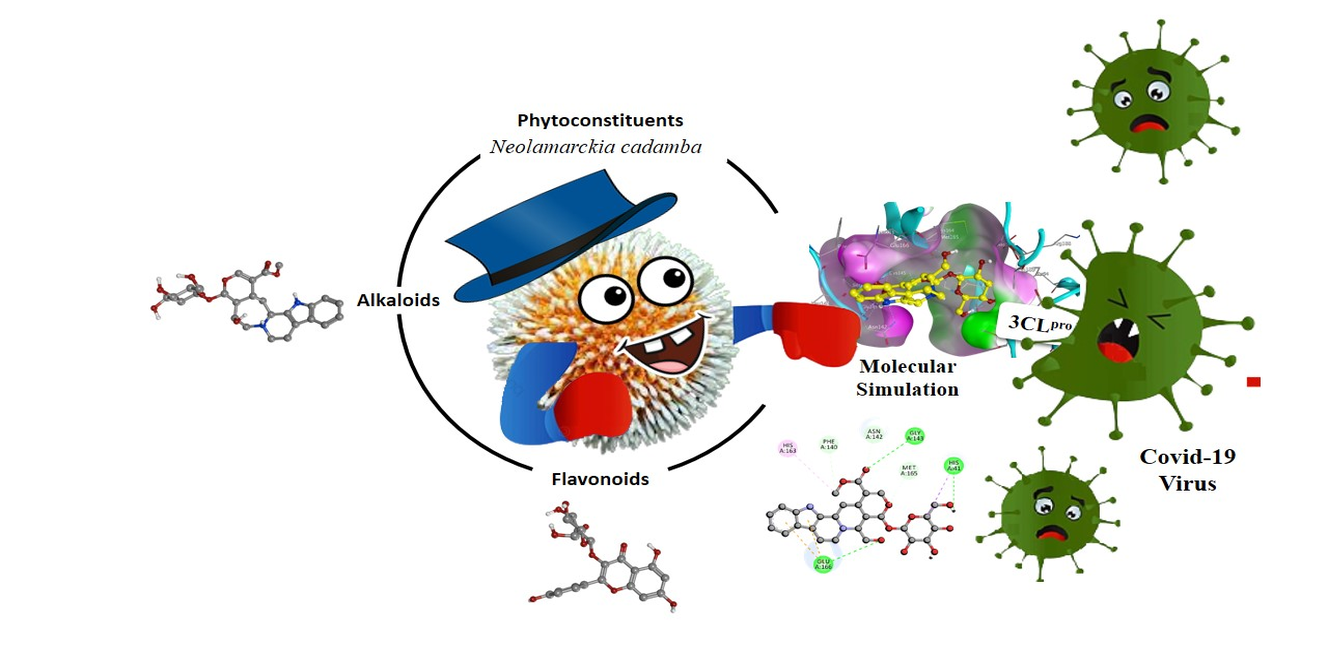
In present study, the inhibitory potential of Neolamarckia cadamba phytoconstituents was investigated against SARS-CoV-2 3CL protease (3CL pro) (PDB ID: 6M2N). Molecular docking was analyzed using AutoDock Vina software by setting the grid parameter as X= -33.163, Y= -65.074 and Z= 41.434 with dimensions of the grid box 25 × 25 × 25 Å. Remdesivir was taken as the standard for comparative analysis along with inhibitor 5, 6, 7-trihydroxy-2-phenyl-4H-chromen-4-one. Furthermore, the exploration of 2 D Hydrogen-bond interactions was performed by Biovia Discovery Studio 4.5 program to identify the interactions between an amino acid of target and ligand followed by assessment of physicochemical properties using Lipinski’s rule and Swiss ADME database. The decent bonding scores of secondary metabolites owing to hydrogen bonding with catalytic residues suggest the effectiveness of these phytochemicals towards 3CLpro. The results are further consolidated positively by Lipinski’s rule and Swiss ADME prediction. Thus reasonably, observations with docking studies suggest possibility of phytochemicals from Neolamarckia cadamba to inhibit the 3CLpro and consequently would be explored further as agents for preventing COVID-19.




